syri
Synteny and Rearrangement Identifier
Plotting genomic structure using plotsr
The output of SyRI can be used with plotsr to generate a graphical represtation of genomic structure using the following command:
python /path/to/plotsr syri.out /path/to/refgenome /path/to/qrygenome
Usage and available parameters:
usage: Arguments for plotting SRs predicted by SyRI [-h] [-s S] [-R] [-f F]
[-H H] [-W W]
[-o {pdf,png,svg}] [-d D]
[-b {agg,cairo,pdf,pgf,ps,svg,template}]
reg r q
positional arguments:
reg syri.out file generated by SyRI
r path to reference genome
q path to query genome
optional arguments:
-h, --help show this help message and exit
-s S minimum size of a SR to be plotted
-R Create ribbons
-f F font size
-H H height of the plot
-W W width of the plot
-o {pdf,png,svg} output file format (pdf, png, svg)
-d D DPI for the final image
-b {agg,cairo,pdf,pgf,ps,svg,template}
Matplotlib backend to use
Example of output plot:
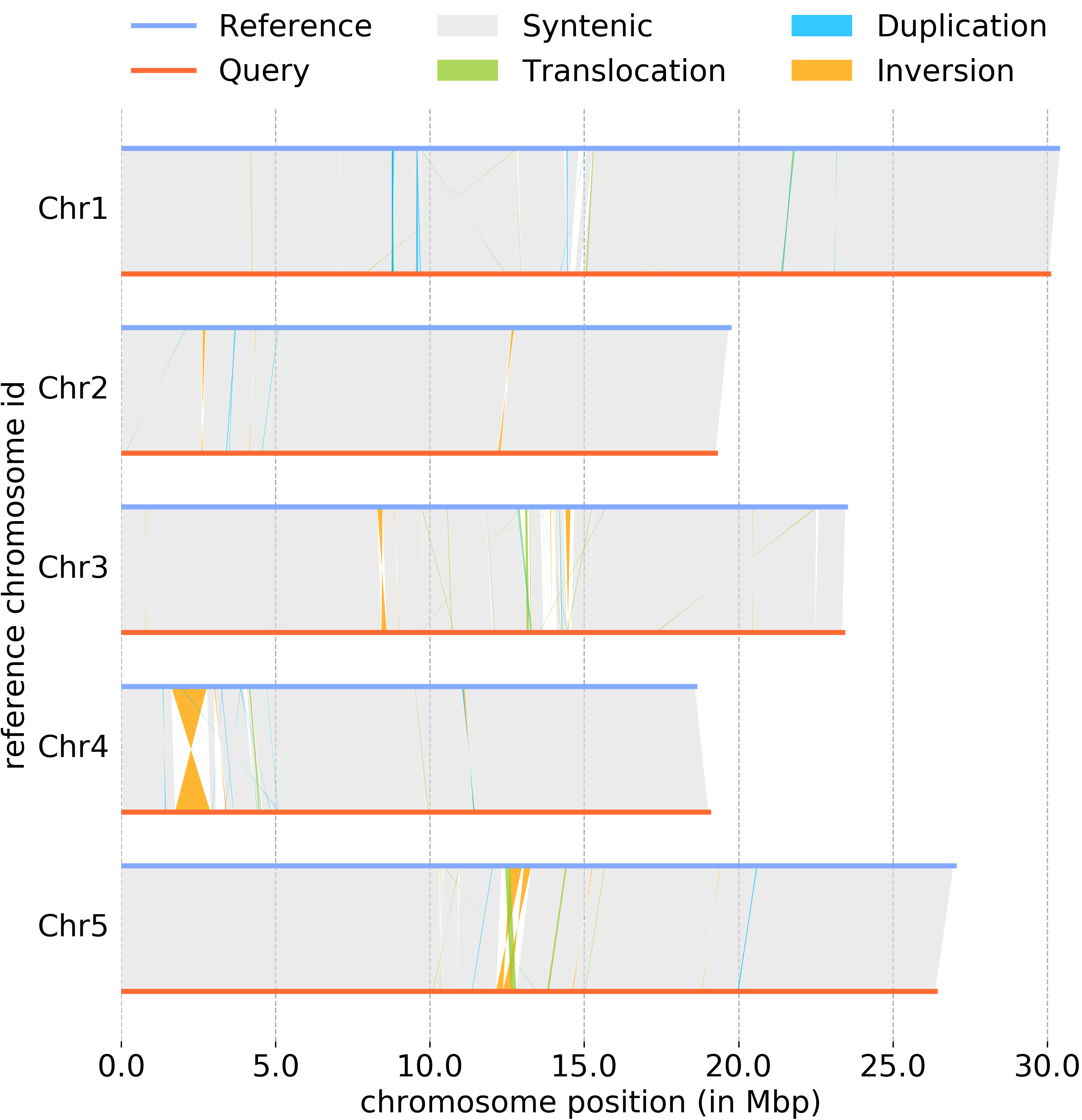
Sample plot generated using plotsr.